Advanced Retrieval Techniques to Supercharge Your RAG
24 May 2025 by dzlabRetrieval Augmented Generation (RAG) has become a cornerstone for building powerful AI applications that can leverage vast amounts of information.
At its heart, RAG relies on vector databases as a knowledge source, storing information as embeddings (i.e. numerical vectors) that represent the semantic meaning of text, images, or other data types in a high-dimensional space. These embeddings are used to query and retrieve relevant information before generating a response from the LLM, improving the model’s ability to provide contextually accurate answers.
While basic vector search with embeddings is a great starting point, we often encounter scenarios where it falls short. It tends to find documents that discuss similar topics as the query, but don’t necessarily contain the direct answer needed. This can lead to the LLM receiving irrelevant information, known as “distractors,” which can degrade the quality of the generated response and make debugging difficult.
This blog post, explores how we can enhance a RAG pipelines using query expansion and re-ranking, with practical examples using ChromaDB and Google’s Generative AI models. The notebook with implementation can be found here.
The Foundation: Embeddings-Based Retrieval
Many teams start with simple retrieval methods, often relying on semantic similarity or basic embeddings. The common workflow involves ingesting documents, splitting them into manageable chunks, embedding these chunks, and storing them in a vector database like ChromaDB. When a user submits a query, this get embed with same way as the chunks were embedded. Then finding documents with the most similar embeddings (nearest neighbors), and feeding those documents as context to the LLM.
Let’s build this basic RAG pipeline:
1. Document Loading & Preprocessing:
We will load a PDF document, and extract its text content. The raw text is then split into smaller, more manageable chunks. We use both RecursiveCharacterTextSplitter and SentenceTransformersTokenTextSplitter to for chunking.
from pypdf import PdfReader
from langchain.text_splitter import RecursiveCharacterTextSplitter, SentenceTransformersTokenTextSplitter
from tqdm import tqdm
filename = 'cisco-annual-report-2023.pdf'
# !curl -s -o {filename} https://www.cisco.com/c/dam/en_us/about/annual-report/{filename} # Download step
reader = PdfReader(filename)
texts = [page.extract_text().strip() for page in reader.pages]
texts = [text for text in texts if text] # filter out empty strings
character_splitter = RecursiveCharacterTextSplitter(
separators=["\n\n", "\n", ". ", " ", ""],
chunk_size=1000,
chunk_overlap=0
)
character_split_texts = character_splitter.split_text('\n\n'.join(texts))
token_splitter = SentenceTransformersTokenTextSplitter(chunk_overlap=0, tokens_per_chunk=256)
token_split_texts = []
for text in tqdm(character_split_texts):
token_split_texts += token_splitter.split_text(text)
2. Embedding & Indexing:
The text chunks are then converted into dense vector embeddings using a sentence transformer model. ChromaDB is used as the vector store, with SentenceTransformerEmbeddingFunction handling the embedding process.
import chromadb
from chromadb.utils.embedding_functions import SentenceTransformerEmbeddingFunction
from pathlib import Path
embedding_function = SentenceTransformerEmbeddingFunction()
chroma_client = chromadb.PersistentClient(path="db/") # Or chromadb.Client() for in-memory
chroma_collection_name = Path(filename).stem
chroma_collection = chroma_client.create_collection(chroma_collection_name, embedding_function=embedding_function)
ids = [str(i) for i in range(len(token_split_texts))]
chroma_collection.add(ids=ids, documents=token_split_texts)
print("Total indexed documents", chroma_collection.count())
3. Retrieval & Generation:
When a user submits a query, we embed it, and ChromaDB is queried to find the most similar document chunks. These retrieved chunks, along with the original query, are then passed to an LLM (like Google’s Gemini) to generate an answer.
import os
import google.generativeai as genai
from functools import lru_cache
import backoff
# from google.api_core.exceptions import InternalServerError, TooManyRequests # For backoff
# genai.configure(api_key=os.environ['GOOGLE_API_KEY']) # Set your API key
@lru_cache(maxsize=128)
def rag(query, topk=5, model_name='models/chat-bison-001'): # model_name can be gemini-pro as well
results = chroma_collection.query(query_texts=[query], n_results=topk)
retrieved_documents = results['documents'][0]
information = '\n\n'.join(retrieved_documents)
if "gemini" in model_name:
model_instance = genai.GenerativeModel(model_name)
response = model_instance.generate_content(f"Question: {query}. \n Information: {information}")
answer = response.text
else: # Assuming PaLM chat model
context = """
You are a helpful expert financial research assistant.
Your users are asking questions about information contained in an annual report.
You will be shown the user's question, and the relevant information from the annual report.
Answer the user's question using only this information.
"""
messages = [{'author': '0', 'content': f"Question: {query}. \n Information: {information}"}]
response = genai.chat(
model=model_name,
context=context,
messages=messages,
temperature=0,
candidate_count=2
)
answer = response.last
return answer
# Example usage:
# answer = rag("What was the total revenue?", model_name='gemini-pro')
# print(answer)
When Simple Vector Search Stumbles
Limitations
While finding documents with similar embeddings seems intuitive, simple vector search based on a general-purpose embedding model isn’t always sufficient for effective RAG. The core issue is that semantic similarity in a high-dimensional embedding space, derived from a model trained on broad language patterns, doesn’t always equate to relevancy for a specific user query or task.
Here’s why simple vector search can stumble:
- Topical Similarity vs. Direct Answers: Embedding models are great at capturing the overall meaning or topic of a document chunk and a query. However, a query might be about a very specific fact or detail. Simple semantic similarity might retrieve documents that talk extensively about the topic but don’t contain the precise piece of information the user needs.
- Lack of Task-Specific Understanding: The embedding model is trained generally and doesn’t inherently understand the specific task the RAG system is trying to accomplish with the retrieved documents (e.g., answering a financial question vs. summarizing a technical paper). The “nearest” neighbors in the general embedding space might not be the most useful for the particular query’s intent.
- The Problem of Distractors: For many queries, especially those that are ambiguous, very general, or completely irrelevant to the document set, simple vector search will still return the “nearest” documents. These results are often irrelevant to the query and are referred to as distractors.
- Impact of Distractors on LLMs: Passing distractors to the LLM as context can significantly degrade the quality of the generated response. The LLM might get “distracted” by the irrelevant information, leading to incorrect, nonsensical, or incomplete answers. Diagnosing and debugging these issues caused by distractors can be challenging.
- Geometric Distribution: Queries can land in different parts of the embedding space relative to the data points. Queries that fall outside dense clusters of relevant information might retrieve documents that are geometrically “nearest” but are spread out and less cohesive in terms of specific relevancy. Conversely, even irrelevant queries will return documents based on proximity in the embedding space, resulting in a context window filled entirely with distractors.
These limitations highlight the need for more sophisticated techniques that can refine the query, re-evaluate the retrieved documents, or adapt the embedding space itself to better align with the specific task and user intent.
Visualizing Embeddings
To better understand the “shape” of the data (user query vs stored documents) and identify potential pitfalls, we can use UMAP (Uniform Manifold Approximation and Projection) to visualize the embeddings in a 2D space. This projection will also help us understand:
- Sparse Regions: Queries might fall into areas of the embedding space where relevant documents are scarce.
- Semantic Ambiguity: A query might be semantically close to irrelevant documents if its embedding isn’t precise enough or if the document embeddings themselves aren’t well-separated.
import umap
import numpy as np
import matplotlib.pyplot as plt
# Assuming 'embeddings' is a list/array of all document embeddings from chroma_collection.get()
umap_transform = umap.UMAP(random_state=0, transform_seed=0).fit(embeddings)
def project_embeddings(embeddings_list, umap_model):
umap_embeddings = np.empty((len(embeddings_list), 2))
for i, embedding in enumerate(tqdm(embeddings_list)):
umap_embeddings[i] = umap_model.transform([embedding])
return umap_embeddings
projected_dataset_embeddings = project_embeddings(embeddings, umap_transform)
# Plotting function
def plot_retrieval(query, projected_dataset_embeddings, umap_transform, chroma_collection, embedding_function):
query_embedding = embedding_function([query])[0]
results = chroma_collection.query(query_texts=[query], n_results=5, include=['embeddings'])
retrieved_embeddings = results['embeddings'][0]
projected_query_embedding = project_embeddings([query_embedding], umap_transform)
projected_retrieved_embeddings = project_embeddings(retrieved_embeddings, umap_transform)
plt.figure()
plt.scatter(projected_dataset_embeddings[:, 0], projected_dataset_embeddings[:, 1], s=10, color='gray', alpha=0.7, label="Dataset document")
plt.scatter(projected_query_embedding[:, 0], projected_query_embedding[:, 1], s=150, marker='x', color='r', label="User Query")
plt.scatter(projected_retrieved_embeddings[:, 0], projected_retrieved_embeddings[:, 1], s=80, facecolors='none', edgecolors='g', label="Retrieved document")
plt.legend()
plt.title(query)
plt.axis('off')
plt.show()
# plot_retrieval("What is the total revenue?", projected_dataset_embeddings, umap_transform, chroma_collection, embedding_function)
The following visualizations highlight why simply picking the top-k nearest neighbors isn’t always optimal. This is obvious when the query has nothig to do with the dataset, but even if it’s relevant the selected documents may not have usefull information for generating a final response.
| Relevant query | Irrelevant query |
|---|---|
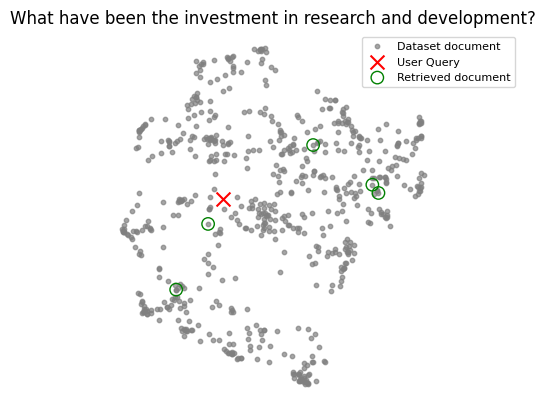 |
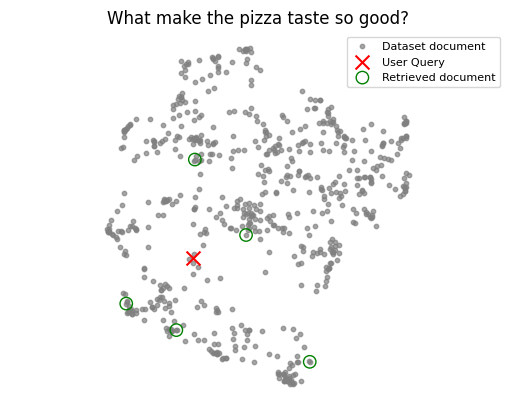 |
Visualizations like these highlight why simply picking the top-k nearest neighbors isn’t always optimal.
Improving the Query Itself: Query Expansion Techniques
One powerful approach is to use an LLM to improve the user’s initial query before sending it to the retrieval system. This section highlights two main techniques for this:
1. Expansion with Generated Answers (HyDE-like approach):
The idea here is to generate a hypothetical answer to the user’s query using an LLM. This hypothetical answer, rich in relevant keywords and concepts, is then concatenated with the original query, creating a richer input. The combined text is embedded and used for retrieval. This often helps bridge the semantic gap between the query and the actual documents. As the retrieval system is guided to find documents that don’t just discuss the topic but actually look like they contain an answer.
@lru_cache(maxsize=128)
def augment_query_generated(query, model_name='models/chat-bison-001'):
context = """
You are a helpful expert financial research assistant.
Provide an example answer to the given question, that might be found in a document like an annual report.
"""
messages = [{'author': '0', 'content': query}]
response = genai.chat(
model=model_name,
context=context,
messages=messages,
temperature=0,
candidate_count=2
)
answer = response.last
return answer
# original_query = "Was there significant turnover in the executive team?"
# hypothetical_answer = augment_query_generated(original_query)
# joint_query = f"{original_query} {hypothetical_answer}"
# results = chroma_collection.query(query_texts=joint_query, n_results=5, include=['documents'])
The following UMAP plots show how this combined input (user query with a synthetic answer) embeddings often shifts closer to relevant document clusters.
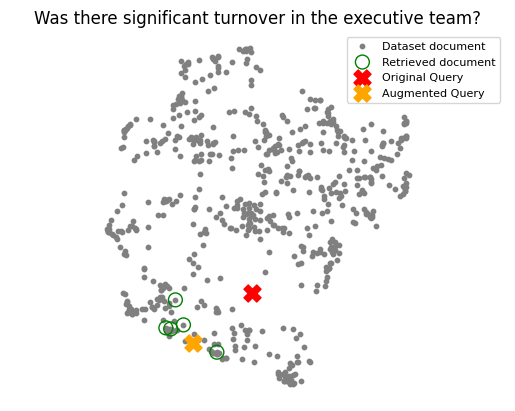
2. Expansion with Multiple Queries:
Instead of one hypothetical answer, we can generate several related questions based on the original query. Each of these (original + augmented queries) is then used to retrieve documents. The results are pooled, deduplicated, and then re-ranked. This expands the search to cover different facets or re-wordings of the user’s need, potentially retrieving relevant information from various parts of the embedding space that a single query might miss.
@lru_cache(maxsize=128)
def augment_multiple_query(query, model_name='models/text-bison-001'): # Using text-bison for this
prompt = f"""
You are a helpful expert financial research assistant. Your users are asking questions about an annual financial report.
Given the user question, suggest up to five additional related questions to help them find the information they need.
The questions should be short without compound sentences and cover different aspects of the topic.
Make sure the questions are complete, and that they are related to the original user question.
Output one question per line. Do not number the questions.
Question: {query}
"""
response = genai.generate_text(
model=model_name,
prompt=prompt,
temperature=0,
candidate_count=2
)
questions = response.candidates[0]['output'].split("\n")
return questions
# original_query = "What were the most important factors that contributed to increases in revenue?"
# augmented_queries = augment_multiple_query(original_query)
# all_queries = [original_query] + augmented_queries
# results = chroma_collection.query(query_texts=all_queries, n_results=5, include=['documents'])
# # ... then deduplicate and re-rank
The following UMAP plots show how this combined input (user query with sub-queries) embeddings often shifts closer to relevant document clusters.
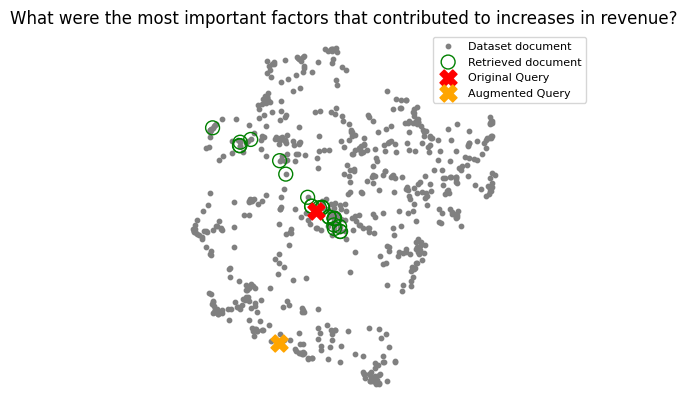
Refining Results: Cross-Encoder Re-ranking
After retrieving a potentially large set of documents (perhaps from query expansion or simply by requesting a longer tail of results from the vector database), the next challenge is to identify and prioritize the most relevant ones for the original query. This is where cross-encoder re-ranking comes in.
Unlike the bi-encoder models (like sentence transformers) used for initial embedding and similarity search (which encode query and documents separately), a cross-encoder takes a pair of inputs – the query and a retrieved document – and outputs a single relevancy score. By applying a cross-encoder to score each retrieved document against the original query, we can re-rank the results, placing the most relevant documents at the top. This is particularly useful after using query expansion, allowing us to filter the combined results from multiple augmented queries down to the most relevant set for the initial user intent.
Cross-encoders are often more computationally intensive than bi-encoders but provide a more nuanced relevancy score because they consider the interaction between the query and the document.
from sentence_transformers import CrossEncoder
cross_encoder = CrossEncoder('cross-encoder/ms-marco-MiniLM-L-6-v2')
# Assuming 'retrieved_documents' is a list of documents from ChromaDB
# and 'original_query' is the user's query
pairs = [[original_query, doc] for doc in retrieved_documents]
scores = cross_encoder.predict(pairs)
# Sort documents by these new scores
new_order = np.argsort(scores)[::-1]
reranked_documents = [retrieved_documents[i] for i in new_order]
Customizing Retrieval with Feedback: Embedding Adaptors
Another advanced technique focuses on directly modifying the query embedding based on feedback, effectively customizing the retrieval system to your specific application or user behavior. A small neural network (or even a linear transformation matrix) inserted into the retrieval pipeline after the initial query embedding but before the similarity search. This lightweight model is trained to adapt the query embeddings to better suit the specific domain of the documents. It uses a training dataset of queries, retrieved documents, and relevancy labels (e.g., +1 for relevant, -1 for irrelevant). The training objective is to adjust the query embedding so that it moves geometrically closer to relevant document embeddings and further away from irrelevant ones in the embedding space. This ‘adapting’ process can stretch or squeeze different dimensions of the embedding vector, emphasizing those most relevant to finding useful results for the types of queries and documents in your specific domain.
This method requires collecting feedback data, which can be generated synthetically using an LLM or, ideally, gathered from actual user interactions with the RAG system.
Here are the steps to implement an Embedding Adaptor:
-
Dataset Creation: A dataset of (query, document, relevance_label) triples is needed. The notebook simulates this by generating queries and then using an LLM (PaLM) to label the relevance of retrieved documents for those queries (1 for relevant, -1 for irrelevant).
-
Adapter Model: A simple linear adapter is a matrix W. The adapted query embedding is
projected_query_embedding = W * query_embedding. -
Training: The goal is to train W such that the cosine similarity between
projected_query_embeddinganddocument_embeddingis high for relevant pairs and low for irrelevant ones. Mean Squared Error (MSE) loss is used.
import torch
from torch.utils.data import TensorDataset
# adapter_query_embeddings, adapter_doc_embeddings, adapter_labels are prepared
dataset = TensorDataset(adapter_query_embeddings, adapter_doc_embeddings, adapter_labels)
mat_size = len(adapter_query_embeddings[0])
adapter_matrix = torch.randn(mat_size, mat_size, requires_grad=True)
def model(query_embedding, document_embedding, weights):
projected_query_embedding = torch.matmul(weights, query_embedding)
predictions = torch.cosine_similarity(projected_query_embedding, document_embedding, dim=0)
return predictions
def mse_loss(predictions, labels):
return torch.nn.MSELoss()(predictions, labels)
# Training loop (simplified)
epochs = 100
lr = 0.01
for epoch in tqdm(range(epochs)):
for query_embedding, document_embedding, label in dataset:
prediction = model(query_embedding, document_embedding, adapter_matrix)
loss = mse_loss(prediction, label)
loss.backward()
with torch.no_grad():
adapter_matrix -= lr * adapter_matrix.grad
adapter_matrix.grad.zero_()
The following UMAP plot shows how the adapter can shift queries into denser, more relevant regions of the embedding space.
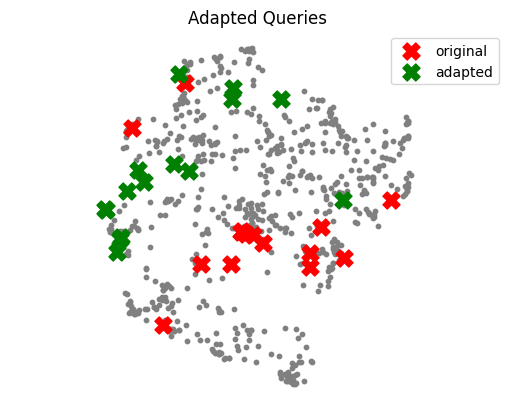
Conclusion
The field of retrieval is rapidly evolving. Beyond simple embeddings-based retrieval techniques, ongoing research explores fine-tuning the base embedding models, fine-tuning the LLM itself to better handle retrieved context, using more complex neural networks for adapters and re-rankers, and developing intelligent methods for optimal document chunking.
In this article we demonstrated that moving beyond basic vector search can significantly improve the quality and relevance of documents retrieved for RAG. Techniques like:
- Query Expansion (HyDE-like and multi-query): Help bridge the semantic gap and retrieve a broader set of potentially relevant documents.
- Cross-Encoder Re-ranking: Provides a more accurate relevance scoring for a candidate set.
- Embedding Adaptors: Offer a powerful way to fine-tune retrieval for specific domains.
By combining these strategies, we can build more robust, accurate, and helpful AI applications, unlocking the full potential of combining large language models with vast amounts of external information.
That’s all folks
I hope you enjoyed this article, feel free to leave a comment or reach out on twitter @bachiirc.